Introduction
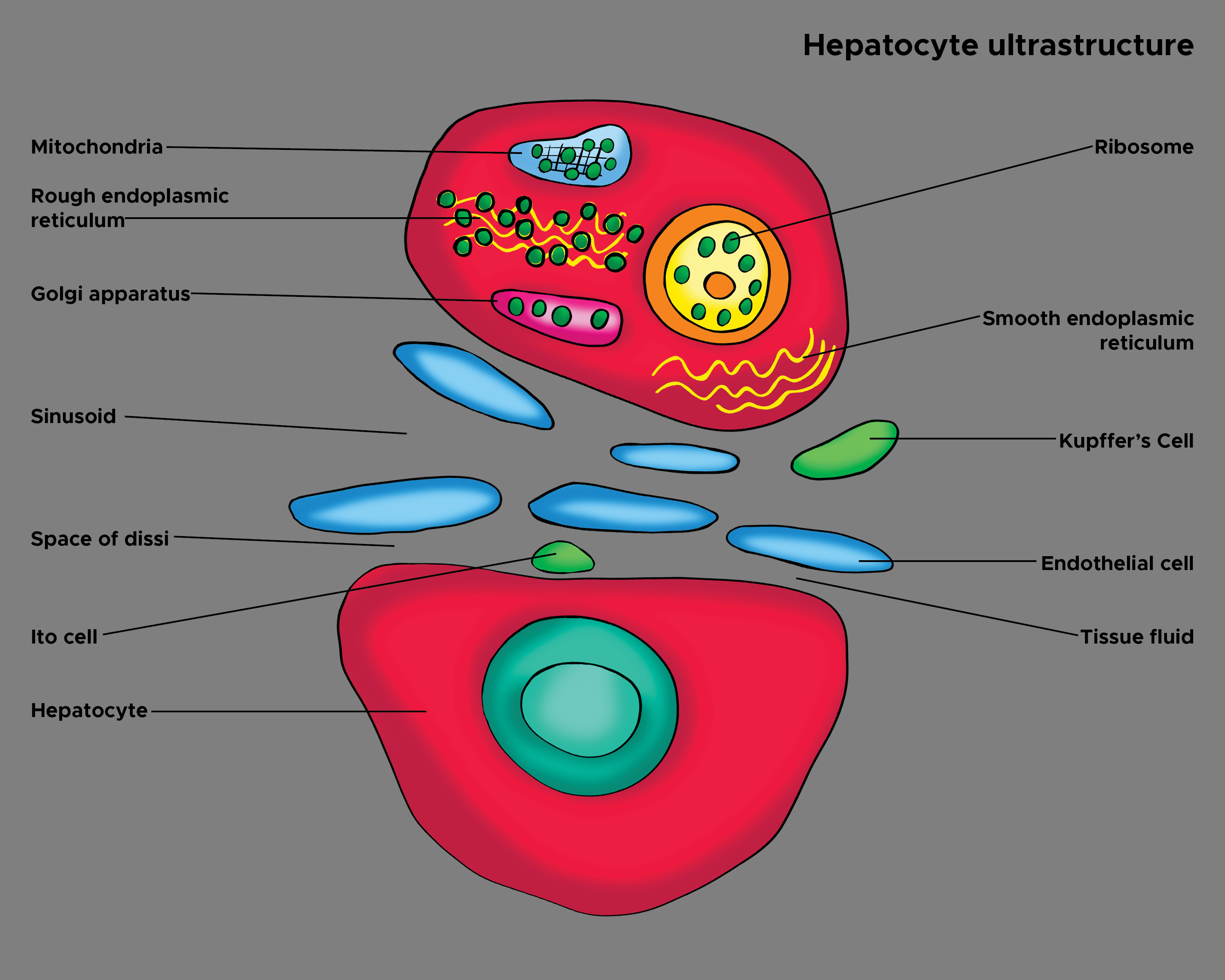
Kupffer cells (also known as stellate sinusoidal macrophages or Kupffer-Browicz cells) are macrophages found in the sinusoids of the liver. (see Image. Hepatocyte Ultrastructure). Kupffer cells make up 80% to 90% of all the macrophages in the entire human body.[1] They are a component of the host immune system and metabolize various compounds. Once thought to be related to endothelial cells, it is now known that the Kupffer cells descend from their macrophage lineage, derived from the yolk sac rather than hematopoietic stem cells.[2]
Differentiation of Kupffer cells is regulated by macrophage colony-stimulating factors (M-CSFs) found in the serum and liver, as well as granulocyte-macrophage colony-stimulating factors (GM-CSFs).[3][4] Kupffer cells can be found in the liver's centrilobular and periportal regions, but they are typically more concentrated in the periportal regions. However, the cells in the 2 regions can differ in certain enzymes, receptors, and subcellular structures.[5]
Structure
Register For Free And Read The Full Article
Search engine and full access to all medical articles
10 free questions in your specialty
Free CME/CE Activities
Free daily question in your email
Save favorite articles to your dashboard
Emails offering discounts
Learn more about a Subscription to StatPearls Point-of-Care
Structure
Kupffer cells are specialized macrophages found in the hepatic sinusoid, along with sinusoidal endothelial cells, Ito cells, and pit cells.[6] Kupffer cells are amoeboid-shaped and are attached to the sinusoidal endothelial cells. [7] Their surface contains microvilli, pseudopodia, and lamellipodia, which can project in all directions. The microvilli and pseudopodia are involved in the endocytosis of particles. They also contain the Golgi apparatus, ribosomes, centrioles, microfilaments, and microtubules in their cytoplasm.[8] Their nucleus is ovoid or indented and can be lobulated. They also contain rough endoplasmic reticulum, nuclear envelope, and annulate lamellae, which all have peroxidase activity.[9]
Kupffer cells' function and structure differ depending on their location in the centrilobular or periportal regions of the liver. Kupffer cells in the periportal regions tend to be larger, have more lysosomal enzyme activities, and have more phagocytic activity, while those in the centrilobular regions produce more superoxide anion.[10]
Function
The lifespan of a Kupffer cell is estimated to be 3.8 days.[11] The primary role of Kupffer cells is to clear foreign debris and particles from the portal system circulation that passes the liver. Kupffer cells can ingest large particles via phagocytosis and small particles and molecules via pinocytosis. It has also been shown that Kupffer cells can migrate to the portal areas and hepatic lymph nodes before they die.[12] In the liver, the population of Kupffer cells is constant, regulated by apoptosis, and phagocytized by neighboring Kupffer cells. In contrast to monocyte-derived macrophages with no proliferative potential, Kupffer cells have a proliferative capacity, allowing regeneration of themselves. In granuloma formation, Kupffer cells are activated without a supply of monocytes, transforming into multinuclear giant cells.[13]
The phagocytic ability of the Kupffer cells is vast; they can engulf pathogens, immune complexes, liposomes, lipid microspheres, tumor cells, endotoxins, and various other particles. Kupffer cells are also known to function heterogeneously based on their location. In zone 1 (periportal) of the liver lobules, they have higher activity overall than their counterpart in zone 3 (centrilobular).[10] The difference in activity is most likely due to the increased exposure to hazardous substances in zone 1 compared to zone 3. In addition to phagocytosis, Kupffer cells can produce inflammatory cytokines, oxygen radicals, TNF-alpha, and proteases; the production of these mediators is thought to contribute to the development of liver injury.[14]
Histochemistry and Cytochemistry
Kupffer cells stain positive for macrophage markers, including ED1, E2, and Ki-M2R in rats and F4/80 in mice. Their lysosomes stain positively for acid phosphatase. Kupffer cells can phagocytize other tracer substances, such as carbon, India ink, or latex microspheres, which are helpful in their identification.[15][16]
Microscopy, Light
Kupffer cells have a wide range of variability in cell size and shape and have elongated cytoplasmic processes. They are found along the sinusoid on top of the endothelium. They can be seen in contact with various cells, such as endothelial cells, fat-storing cells, collagen fibers, and other Kupffer cells.[6]
Microscopy, Electron
On electron microscopy, Kupffer cells are adjacent to the sinusoid but not directly attached to the basement membrane. The microvilli from the parenchymal cells and the pseudopods of the Kupffer cells are intertwined.[8]
Pathophysiology
Kupffer cells are involved in the pathogenesis of liver injury in response to sepsis. The liver macrophages release IL-1 and TNF-alpha, activating leukocytes and sinusoidal endothelial cells to express ICAM-1.[14] The result is tissue damage to the endothelium due to oxygen radicals, proteases, prostanoids, and other substances from leukocytes.[17]
Clinical Significance
Kupffer cells contain the SR-AI/II scavenger receptor, which recognizes and binds the lipid A domain of lipopolysaccharide (LPS) and lipoteichoic acid.[18] Lipopolysaccharide (LPS) is a bacterial endotoxin found in the cell wall of gram-negative bacteria, while lipoteichoic acid is found in gram-positive bacteria. Studies have found that mice lacking SR-AI/II receptors are more susceptible to infection from gram-positive bacteria, suggesting the importance of Kupffer cells in removing bacterial toxins from the system.[19]
Kupffer cells have been identified to play a key role in developing alcohol-induced liver disease. The intestinal tract of humans contains numerous bacteria, which can lead to the production of gut-derived endotoxin. The gut-derived endotoxins make their way to the liver and are cleared by the Kupffer cells. Studies suggest that these endotoxins activate Kupffer cells.[20] Several mechanisms have been proposed regarding the connection between endotoxin levels and alcoholic consumption. One example is that chronic alcohol use prevents Kupffer cells from effectively removing endotoxins from the blood, leading to increased circulating levels of endotoxins. Another mechanism states that alcohol consumption can lead to increased intestinal absorption of endotoxins through increased gut permeability. The endotoxin interacts with both Toll-like receptor 4 (TLR4) and CD14 receptors on Kupffer cells, signaling for the internalization of the lipopolysaccharide (LPS) endotoxin.[21]
Kupffer cell activation produces reactive oxygen species (ROS), such as superoxide, leading to oxidative stress in the liver. TLR4 activates interleukin-1 receptor-associated kinase (IRAK-1), which leads to the activation of nuclear factor kappa B (NF-kB).[20] The activation leads to numerous responses, which include the generation of superoxide and the production of cytokines; the result is liver damage and, eventually, loss of liver function. Kupffer cell activation has also been implicated in developing binge drinking-induced fatty liver disease. The process is mediated by TNF-alpha activation of lipolysis. Kupffer cells can activate inflammasomes, which trigger the activation caspase-1 and the production of IL-1beta, a pro-inflammatory mediator in alcoholic liver disease. Potential treatments for alcoholic liver disease are aimed at Kupffer cells’ role in the pathogenesis of the disease. Treatments for suppressing Kupffer cell activation and eliminating Kupffer cell cytotoxic products include antibiotics, probiotics, TNF-alpha, and IL-1beta inhibitors.[22]
Kupffer cells have Fc, C3, and scavenger receptors involved in the phagocytosis of opsonized and nonopsonized materials.[23] Scavenger receptors are also involved in the deposition of cholesterol in arterial walls, leading to atherogenesis.[24] Also, Kupffer cells remove aged erythrocytes from circulation, resulting in elevated levels of heme oxygenase shortly after phagocytosis. Heme oxygenase is part of the metabolism and production of bilirubin; the enzyme degrades heme molecules in erythrocytes.[25]
Media
(Click Image to Enlarge)
References
Chaudhry S, Emond J, Griesemer A. Immune Cell Trafficking to the Liver. Transplantation. 2019 Jul:103(7):1323-1337. doi: 10.1097/TP.0000000000002690. Epub [PubMed PMID: 30817405]
Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, Rodewald HR. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015 Feb 26:518(7540):547-51. doi: 10.1038/nature13989. Epub 2014 Dec 3 [PubMed PMID: 25470051]
Level 3 (low-level) evidenceYamamoto T, Kaizu C, Kawasaki T, Hasegawa G, Umezu H, Ohashi R, Sakurada J, Jiang S, Shultz L, Naito M. Macrophage colony-stimulating factor is indispensable for repopulation and differentiation of Kupffer cells but not for splenic red pulp macrophages in osteopetrotic (op/op) mice after macrophage depletion. Cell and tissue research. 2008 May:332(2):245-56. doi: 10.1007/s00441-008-0586-8. Epub 2008 Mar 12 [PubMed PMID: 18335245]
Level 3 (low-level) evidenceLi W,He F, Infusion of Kupffer Cells Expanded in {i}Vitro{/i} Ameliorated Liver Fibrosis in a Murine Model of Liver Injury. Cell transplantation. 2021 Jan-Dec [PubMed PMID: 33784833]
Kolios G, Valatas V, Kouroumalis E. Role of Kupffer cells in the pathogenesis of liver disease. World journal of gastroenterology. 2006 Dec 14:12(46):7413-20 [PubMed PMID: 17167827]
Level 3 (low-level) evidenceWisse E, Braet F, Luo D, De Zanger R, Jans D, Crabbé E, Vermoesen A. Structure and function of sinusoidal lining cells in the liver. Toxicologic pathology. 1996 Jan-Feb:24(1):100-11 [PubMed PMID: 8839287]
Level 3 (low-level) evidenceNaito M, Hasegawa G, Ebe Y, Yamamoto T. Differentiation and function of Kupffer cells. Medical electron microscopy : official journal of the Clinical Electron Microscopy Society of Japan. 2004 Mar:37(1):16-28 [PubMed PMID: 15057601]
Level 3 (low-level) evidenceSichel G,Scalia M,Corsaro C, Amphibia Kupffer cells. Microscopy research and technique. 2002 Jun 15 [PubMed PMID: 12112430]
Level 3 (low-level) evidenceWisse E. Observations on the fine structure and peroxidase cytochemistry of normal rat liver Kupffer cells. Journal of ultrastructure research. 1974 Mar:46(3):393-426 [PubMed PMID: 4363811]
Level 3 (low-level) evidenceCampion SN, Tatis-Rios C, Augustine LM, Goedken MJ, van Rooijen N, Cherrington NJ, Manautou JE. Effect of allyl alcohol on hepatic transporter expression: zonal patterns of expression and role of Kupffer cell function. Toxicology and applied pharmacology. 2009 Apr 1:236(1):49-58. doi: 10.1016/j.taap.2009.01.007. Epub 2009 Jan 24 [PubMed PMID: 19371622]
Level 3 (low-level) evidenceNguyen-Lefebvre AT, Horuzsko A. Kupffer Cell Metabolism and Function. Journal of enzymology and metabolism. 2015:1(1):. pii: 101. Epub 2015 Aug 14 [PubMed PMID: 26937490]
Hardonk MJ,Dijkhuis FW,Grond J,Koudstaal J,Poppema S, Evidence for a migratory capability of rat Kupffer cells to portal tracts and hepatic lymph nodes. Virchows Archiv. B, Cell pathology including molecular pathology. 1986 [PubMed PMID: 2876547]
Level 3 (low-level) evidenceYamada M, Naito M, Takahashi K. Kupffer cell proliferation and glucan-induced granuloma formation in mice depleted of blood monocytes by strontium-89. Journal of leukocyte biology. 1990 Mar:47(3):195-205 [PubMed PMID: 2307905]
Level 3 (low-level) evidenceRoberts RA, Ganey PE, Ju C, Kamendulis LM, Rusyn I, Klaunig JE. Role of the Kupffer cell in mediating hepatic toxicity and carcinogenesis. Toxicological sciences : an official journal of the Society of Toxicology. 2007 Mar:96(1):2-15 [PubMed PMID: 17122412]
Level 3 (low-level) evidenceElchaninov AV, Fatkhudinov TK, Vishnyakova PA, Lokhonina AV, Sukhikh GT. Phenotypical and Functional Polymorphism of Liver Resident Macrophages. Cells. 2019 Sep 5:8(9):. doi: 10.3390/cells8091032. Epub 2019 Sep 5 [PubMed PMID: 31491903]
Fujita H, Kawamata S, Yamashita K. Electron microscopic studies on multinucleate foreign body giant cells derived from Kupffer cells in mice given Indian ink intravenously. Virchows Archiv. B, Cell pathology including molecular pathology. 1983:42(1):33-42 [PubMed PMID: 6132487]
Level 3 (low-level) evidenceGulubova MV. Intercellular adhesion molecule-1 (ICAM-1) expression in the liver of patients with extrahepatic cholestasis. Acta histochemica. 1998 Feb:100(1):59-74 [PubMed PMID: 9542581]
van Oosten M, van Amersfoort ES, van Berkel TJ, Kuiper J. Scavenger receptor-like receptors for the binding of lipopolysaccharide and lipoteichoic acid to liver endothelial and Kupffer cells. Journal of endotoxin research. 2001:7(5):381-4 [PubMed PMID: 11753207]
Level 3 (low-level) evidenceThomas CA, Li Y, Kodama T, Suzuki H, Silverstein SC, El Khoury J. Protection from lethal gram-positive infection by macrophage scavenger receptor-dependent phagocytosis. The Journal of experimental medicine. 2000 Jan 3:191(1):147-56 [PubMed PMID: 10620613]
Level 3 (low-level) evidenceLuedde T, Schwabe RF. NF-κB in the liver--linking injury, fibrosis and hepatocellular carcinoma. Nature reviews. Gastroenterology & hepatology. 2011 Feb:8(2):108-18. doi: 10.1038/nrgastro.2010.213. Epub [PubMed PMID: 21293511]
Ciesielska A, Matyjek M, Kwiatkowska K. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cellular and molecular life sciences : CMLS. 2021 Feb:78(4):1233-1261. doi: 10.1007/s00018-020-03656-y. Epub 2020 Oct 15 [PubMed PMID: 33057840]
Chen W, Zhang J, Fan HN, Zhu JS. Function and therapeutic advances of chemokine and its receptor in nonalcoholic fatty liver disease. Therapeutic advances in gastroenterology. 2018:11():1756284818815184. doi: 10.1177/1756284818815184. Epub 2018 Dec 6 [PubMed PMID: 30574191]
Level 3 (low-level) evidenceBilzer M, Roggel F, Gerbes AL. Role of Kupffer cells in host defense and liver disease. Liver international : official journal of the International Association for the Study of the Liver. 2006 Dec:26(10):1175-86 [PubMed PMID: 17105582]
Level 3 (low-level) evidencevan Oosten M, van de Bilt E, van Berkel TJ, Kuiper J. New scavenger receptor-like receptors for the binding of lipopolysaccharide to liver endothelial and Kupffer cells. Infection and immunity. 1998 Nov:66(11):5107-12 [PubMed PMID: 9784510]
Level 3 (low-level) evidenceHirano K, Kobayashi T, Watanabe T, Yamamoto T, Hasegawa G, Hatakeyama K, Suematsu M, Naito M. Role of heme oxygenase-1 and Kupffer cells in the production of bilirubin in the rat liver. Archives of histology and cytology. 2001 May:64(2):169-78 [PubMed PMID: 11436987]
Level 3 (low-level) evidence