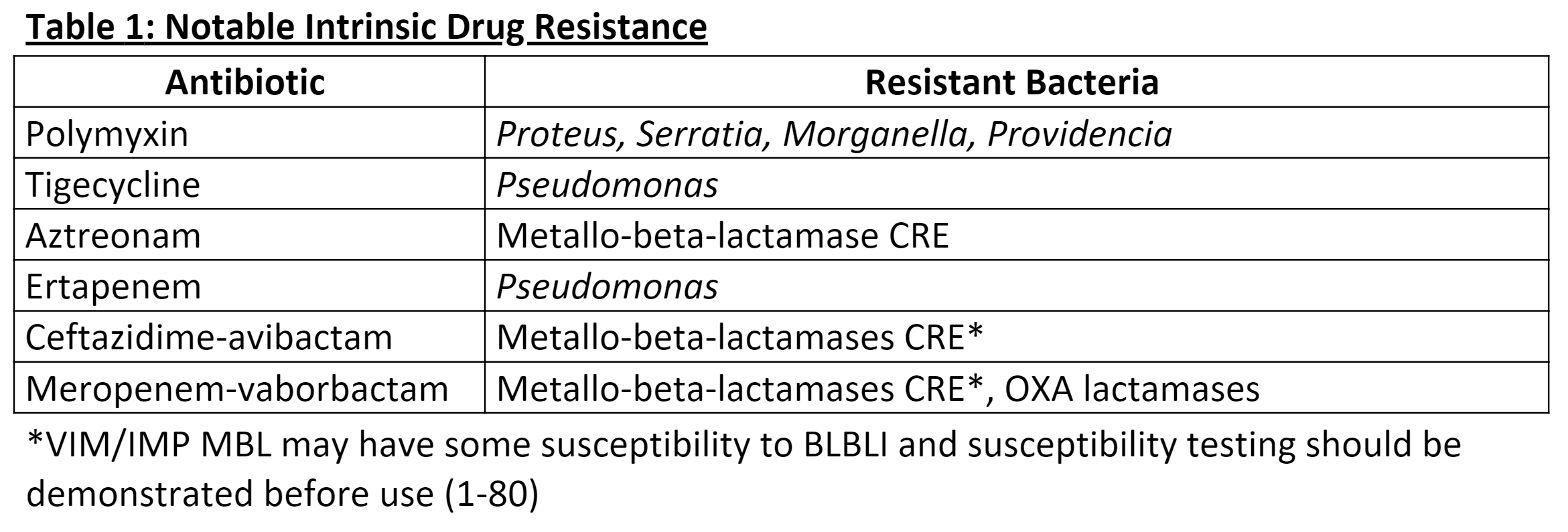
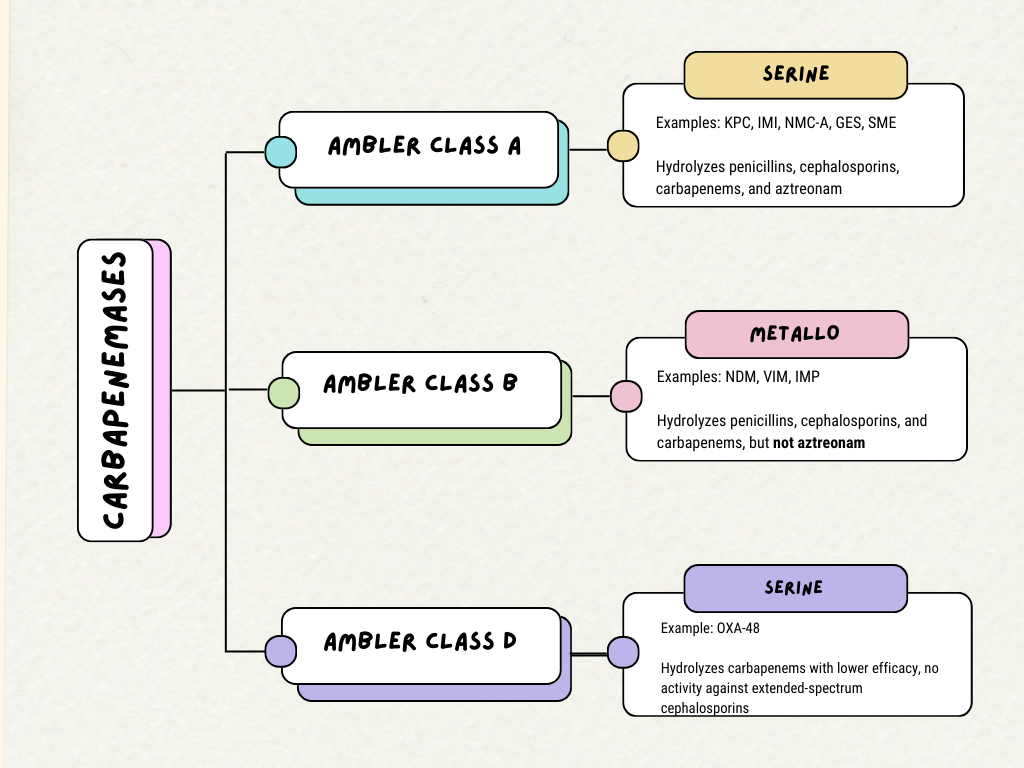
[1]
van Duin D, Paterson DL. Multidrug-Resistant Bacteria in the Community: An Update. Infectious disease clinics of North America. 2020 Dec:34(4):709-722. doi: 10.1016/j.idc.2020.08.002. Epub 2020 Sep 30
[PubMed PMID: 33011046]
[2]
Darby EM, Trampari E, Siasat P, Gaya MS, Alav I, Webber MA, Blair JMA. Molecular mechanisms of antibiotic resistance revisited. Nature reviews. Microbiology. 2023 May:21(5):280-295. doi: 10.1038/s41579-022-00820-y. Epub 2022 Nov 21
[PubMed PMID: 36411397]
[3]
Al-Tawfiq JA, Momattin H, Al-Ali AY, Eljaaly K, Tirupathi R, Haradwala MB, Areti S, Alhumaid S, Rabaan AA, Al Mutair A, Schlagenhauf P. Antibiotics in the pipeline: a literature review (2017-2020). Infection. 2022 Jun:50(3):553-564. doi: 10.1007/s15010-021-01709-3. Epub 2021 Oct 4
[PubMed PMID: 34606056]
[4]
Munita JM, Arias CA. Mechanisms of Antibiotic Resistance. Microbiology spectrum. 2016 Apr:4(2):. doi: 10.1128/microbiolspec.VMBF-0016-2015. Epub
[PubMed PMID: 27227291]
[5]
Jacoby GA. AmpC beta-lactamases. Clinical microbiology reviews. 2009 Jan:22(1):161-82, Table of Contents. doi: 10.1128/CMR.00036-08. Epub
[PubMed PMID: 19136439]
[6]
Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D, Clancy CJ. Infectious Diseases Society of America 2023 Guidance on the Treatment of Antimicrobial Resistant Gram-Negative Infections. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2023 Jul 18:():. pii: ciad428. doi: 10.1093/cid/ciad428. Epub 2023 Jul 18
[PubMed PMID: 37463564]
[7]
Vardakas KZ, Tansarli GS, Rafailidis PI, Falagas ME. Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to Enterobacteriaceae producing extended-spectrum β-lactamases: a systematic review and meta-analysis. The Journal of antimicrobial chemotherapy. 2012 Dec:67(12):2793-803. doi: 10.1093/jac/dks301. Epub 2012 Aug 21
[PubMed PMID: 22915465]
Level 1 (high-level) evidence
[8]
De Rosa M, Verdino A, Soriente A, Marabotti A. The Odd Couple(s): An Overview of Beta-Lactam Antibiotics Bearing More Than One Pharmacophoric Group. International journal of molecular sciences. 2021 Jan 9:22(2):. doi: 10.3390/ijms22020617. Epub 2021 Jan 9
[PubMed PMID: 33435500]
Level 3 (low-level) evidence
[9]
Wong D, Spellberg B. Leveraging antimicrobial stewardship into improving rates of carbapenem-resistant Enterobacteriaceae. Virulence. 2017 May 19:8(4):383-390. doi: 10.1080/21505594.2016.1188234. Epub 2016 May 17
[PubMed PMID: 27187821]
[10]
Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clinical microbiology reviews. 2012 Oct:25(4):682-707. doi: 10.1128/CMR.05035-11. Epub
[PubMed PMID: 23034326]
[11]
Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. The Lancet. Infectious diseases. 2013 Sep:13(9):785-96. doi: 10.1016/S1473-3099(13)70190-7. Epub
[PubMed PMID: 23969216]
[12]
Codjoe FS, Donkor ES. Carbapenem Resistance: A Review. Medical sciences (Basel, Switzerland). 2017 Dec 21:6(1):. doi: 10.3390/medsci6010001. Epub 2017 Dec 21
[PubMed PMID: 29267233]
[13]
Hammoudi Halat D, Ayoub Moubareck C. The Current Burden of Carbapenemases: Review of Significant Properties and Dissemination among Gram-Negative Bacteria. Antibiotics (Basel, Switzerland). 2020 Apr 16:9(4):. doi: 10.3390/antibiotics9040186. Epub 2020 Apr 16
[PubMed PMID: 32316342]
[14]
Rabaan AA, Eljaaly K, Alhumaid S, Albayat H, Al-Adsani W, Sabour AA, Alshiekheid MA, Al-Jishi JM, Khamis F, Alwarthan S, Alhajri M, Alfaraj AH, Tombuloglu H, Garout M, Alabdullah DM, Mohammed EAE, Yami FSA, Almuhtaresh HA, Livias KA, Mutair AA, Almushrif SA, Abusalah MAHA, Ahmed N. An Overview on Phenotypic and Genotypic Characterisation of Carbapenem-Resistant Enterobacterales. Medicina (Kaunas, Lithuania). 2022 Nov 19:58(11):. doi: 10.3390/medicina58111675. Epub 2022 Nov 19
[PubMed PMID: 36422214]
Level 3 (low-level) evidence
[15]
Marsik FJ, Nambiar S. Review of carbapenemases and AmpC-beta lactamases. The Pediatric infectious disease journal. 2011 Dec:30(12):1094-5. doi: 10.1097/INF.0b013e31823c0e47. Epub
[PubMed PMID: 22105420]
[16]
Boyd SE, Livermore DM, Hooper DC, Hope WW. Metallo-β-Lactamases: Structure, Function, Epidemiology, Treatment Options, and the Development Pipeline. Antimicrobial agents and chemotherapy. 2020 Sep 21:64(10):. doi: 10.1128/AAC.00397-20. Epub 2020 Sep 21
[PubMed PMID: 32690645]
[17]
Hirvonen VHA, Spencer J, van der Kamp MW. Antimicrobial Resistance Conferred by OXA-48 β-Lactamases: Towards a Detailed Mechanistic Understanding. Antimicrobial agents and chemotherapy. 2021 May 18:65(6):. doi: 10.1128/AAC.00184-21. Epub 2021 May 18
[PubMed PMID: 33753332]
Level 3 (low-level) evidence
[18]
van Duin D, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. 2017 May 19:8(4):460-469. doi: 10.1080/21505594.2016.1222343. Epub 2016 Aug 11
[PubMed PMID: 27593176]
[19]
Cui X, Zhang H, Du H. Carbapenemases in Enterobacteriaceae: Detection and Antimicrobial Therapy. Frontiers in microbiology. 2019:10():1823. doi: 10.3389/fmicb.2019.01823. Epub 2019 Aug 20
[PubMed PMID: 31481937]
[20]
Livermore DM, Nicolau DP, Hopkins KL, Meunier D. Carbapenem-Resistant Enterobacterales, Carbapenem Resistant Organisms, Carbapenemase-Producing Enterobacterales, and Carbapenemase-Producing Organisms: Terminology Past its "Sell-By Date" in an Era of New Antibiotics and Regional Carbapenemase Epidemiology. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2020 Oct 23:71(7):1776-1782. doi: 10.1093/cid/ciaa122. Epub
[PubMed PMID: 32025698]
[21]
Poirel L, Benouda A, Hays C, Nordmann P. Emergence of NDM-1-producing Klebsiella pneumoniae in Morocco. The Journal of antimicrobial chemotherapy. 2011 Dec:66(12):2781-3. doi: 10.1093/jac/dkr384. Epub 2011 Sep 19
[PubMed PMID: 21930570]
[22]
Potter RF, D'Souza AW, Dantas G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2016 Nov:29():30-46. doi: 10.1016/j.drup.2016.09.002. Epub 2016 Sep 19
[PubMed PMID: 27912842]
Level 3 (low-level) evidence
[23]
Haeili M, Barmudeh S, Omrani M, Zeinalzadeh N, Kafil HS, Batignani V, Ghodousi A, Cirillo DM. Whole-genome sequence analysis of clinically isolated carbapenem resistant Escherichia coli from Iran. BMC microbiology. 2023 Feb 27:23(1):49. doi: 10.1186/s12866-023-02796-y. Epub 2023 Feb 27
[PubMed PMID: 36850019]
[24]
Arnold RS, Thom KA, Sharma S, Phillips M, Kristie Johnson J, Morgan DJ. Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. Southern medical journal. 2011 Jan:104(1):40-5. doi: 10.1097/SMJ.0b013e3181fd7d5a. Epub
[PubMed PMID: 21119555]
[25]
Salamzade R, Manson AL, Walker BJ, Brennan-Krohn T, Worby CJ, Ma P, He LL, Shea TP, Qu J, Chapman SB, Howe W, Young SK, Wurster JI, Delaney ML, Kanjilal S, Onderdonk AB, Bittencourt CE, Gussin GM, Kim D, Peterson EM, Ferraro MJ, Hooper DC, Shenoy ES, Cuomo CA, Cosimi LA, Huang SS, Kirby JE, Pierce VM, Bhattacharyya RP, Earl AM. Inter-species geographic signatures for tracing horizontal gene transfer and long-term persistence of carbapenem resistance. Genome medicine. 2022 Apr 5:14(1):37. doi: 10.1186/s13073-022-01040-y. Epub 2022 Apr 5
[PubMed PMID: 35379360]
[27]
Tooke CL, Hinchliffe P, Bragginton EC, Colenso CK, Hirvonen VHA, Takebayashi Y, Spencer J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. Journal of molecular biology. 2019 Aug 23:431(18):3472-3500. doi: 10.1016/j.jmb.2019.04.002. Epub 2019 Apr 5
[PubMed PMID: 30959050]
[28]
Gustaferro CA, Steckelberg JM. Cephalosporin antimicrobial agents and related compounds. Mayo Clinic proceedings. 1991 Oct:66(10):1064-73
[PubMed PMID: 1921490]
[29]
Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. Carbapenems: past, present, and future. Antimicrobial agents and chemotherapy. 2011 Nov:55(11):4943-60. doi: 10.1128/AAC.00296-11. Epub 2011 Aug 22
[PubMed PMID: 21859938]
[30]
Jeon JH, Lee JH, Lee JJ, Park KS, Karim AM, Lee CR, Jeong BC, Lee SH. Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. International journal of molecular sciences. 2015 Apr 29:16(5):9654-92. doi: 10.3390/ijms16059654. Epub 2015 Apr 29
[PubMed PMID: 25938965]
[31]
Lepe JA, Martínez-Martínez L. Resistance mechanisms in Gram-negative bacteria. Medicina intensiva. 2022 Jul:46(7):392-402. doi: 10.1016/j.medine.2022.05.004. Epub 2022 May 31
[PubMed PMID: 35660283]
[32]
Franco MR, Caiaffa-Filho HH, Burattini MN, Rossi F. Metallo-beta-lactamases among imipenem-resistant Pseudomonas aeruginosa in a Brazilian university hospital. Clinics (Sao Paulo, Brazil). 2010:65(9):825-9
[PubMed PMID: 21049207]
[33]
Sotgiu G, Are BM, Pesapane L, Palmieri A, Muresu N, Cossu A, Dettori M, Azara A, Mura II, Cocuzza C, Aliberti S, Piana A. Nosocomial transmission of carbapenem-resistant Klebsiella pneumoniae in an Italian university hospital: a molecular epidemiological study. The Journal of hospital infection. 2018 Aug:99(4):413-418. doi: 10.1016/j.jhin.2018.03.033. Epub 2018 Apr 3
[PubMed PMID: 29621600]
Level 2 (mid-level) evidence
[34]
Doumith M, Ellington MJ, Livermore DM, Woodford N. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. The Journal of antimicrobial chemotherapy. 2009 Apr:63(4):659-67. doi: 10.1093/jac/dkp029. Epub 2009 Feb 20
[PubMed PMID: 19233898]
[35]
Chen HY, Jean SS, Lee YL, Lu MC, Ko WC, Liu PY, Hsueh PR. Carbapenem-Resistant Enterobacterales in Long-Term Care Facilities: A Global and Narrative Review. Frontiers in cellular and infection microbiology. 2021:11():601968. doi: 10.3389/fcimb.2021.601968. Epub 2021 Apr 23
[PubMed PMID: 33968793]
Level 3 (low-level) evidence
[36]
Pérez-Galera S, Bravo-Ferrer JM, Paniagua M, Kostyanev T, de Kraker MEA, Feifel J, Sojo-Dorado J, Schotsman J, Cantón R, Daikos GL, Carevic B, Dragovac G, Tan LK, Raka L, Hristea A, Viale P, Akova M, Reguera JM, Valiente de Santis L, Torre-Cisneros J, Cano Á, Roilides E, Radulovic L, Kirakli C, Shaw E, Falagas ME, Pintado V, Goossens H, Bonten MJ, Gutiérrez-Gutiérrez B, Rodriguez-Baño J, COMBACTE-CARE-EURECA Team. Risk factors for infections caused by carbapenem-resistant Enterobacterales: an international matched case-control-control study (EURECA). EClinicalMedicine. 2023 Mar:57():101871. doi: 10.1016/j.eclinm.2023.101871. Epub 2023 Feb 27
[PubMed PMID: 36895801]
Level 2 (mid-level) evidence
[37]
Zeng M, Xia J, Zong Z, Shi Y, Ni Y, Hu F, Chen Y, Zhuo C, Hu B, Lv X, Li J, Liu Z, Zhang J, Yang W, Yang F, Yang Q, Zhou H, Li X, Wang J, Li Y, Ren J, Chen B, Chen D, Wu A, Guan X, Qu J, Wu D, Huang X, Qiu H, Xu Y, Yu Y, Wang M, Society of Bacterial Infection and Resistance of Chinese Medical Association, Expert Committee on Clinical Use of Antimicrobial Agents and Evaluation of Antimicrobial Resistance of the National Health Commission, Infectious Diseases Society of Chinese Medical Education Association. Guidelines for the diagnosis, treatment, prevention and control of infections caused by carbapenem-resistant gram-negative bacilli. Journal of microbiology, immunology, and infection = Wei mian yu gan ran za zhi. 2023 Aug:56(4):653-671. doi: 10.1016/j.jmii.2023.01.017. Epub 2023 Feb 18
[PubMed PMID: 36868960]
Level 2 (mid-level) evidence
[38]
Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against β-lactam antibiotics. Journal of clinical microbiology. 2012 Mar:50(3):927-37. doi: 10.1128/JCM.05737-11. Epub 2012 Jan 11
[PubMed PMID: 22205812]
[39]
Rodríguez-Baño J, Cisneros JM, Gudiol C, Martínez JA. Treatment of infections caused by carbapenemase-producing Enterobacteriaceae. Enfermedades infecciosas y microbiologia clinica. 2014 Dec:32 Suppl 4():49-55. doi: 10.1016/S0213-005X(14)70174-0. Epub
[PubMed PMID: 25542052]
[40]
Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D, Clancy CJ. Infectious Diseases Society of America 2022 Guidance on the Treatment of Extended-Spectrum β-lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa). Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2022 Aug 25:75(2):187-212. doi: 10.1093/cid/ciac268. Epub
[PubMed PMID: 35439291]
[41]
Kazmierczak KM, Tsuji M, Wise MG, Hackel M, Yamano Y, Echols R, Sahm DF. In vitro activity of cefiderocol, a siderophore cephalosporin, against a recent collection of clinically relevant carbapenem-non-susceptible Gram-negative bacilli, including serine carbapenemase- and metallo-β-lactamase-producing isolates (SIDERO-WT-2014 Study). International journal of antimicrobial agents. 2019 Feb:53(2):177-184. doi: 10.1016/j.ijantimicag.2018.10.007. Epub 2018 Oct 26
[PubMed PMID: 30395986]
[42]
Yahav D, Lador A, Paul M, Leibovici L. Efficacy and safety of tigecycline: a systematic review and meta-analysis. The Journal of antimicrobial chemotherapy. 2011 Sep:66(9):1963-71. doi: 10.1093/jac/dkr242. Epub 2011 Jun 18
[PubMed PMID: 21685488]
Level 1 (high-level) evidence
[43]
Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Therapeutic advances in infectious disease. 2016 Feb:3(1):15-21. doi: 10.1177/2049936115621709. Epub
[PubMed PMID: 26862399]
Level 3 (low-level) evidence
[44]
Doi Y, Wachino JI, Arakawa Y. Aminoglycoside Resistance: The Emergence of Acquired 16S Ribosomal RNA Methyltransferases. Infectious disease clinics of North America. 2016 Jun:30(2):523-537. doi: 10.1016/j.idc.2016.02.011. Epub
[PubMed PMID: 27208771]
[45]
Li JJ, Sheng ZK, Deng M, Bi S, Hu FS, Miao HF, Ji ZK, Sheng JF, Li LJ. Epidemic of Klebsiella pneumoniae ST11 clone coproducing KPC-2 and 16S rRNA methylase RmtB in a Chinese University Hospital. BMC infectious diseases. 2012 Dec 23:12():373. doi: 10.1186/1471-2334-12-373. Epub 2012 Dec 23
[PubMed PMID: 23259910]
[46]
Karageorgopoulos DE, Miriagou V, Tzouvelekis LS, Spyridopoulou K, Daikos GL. Emergence of resistance to fosfomycin used as adjunct therapy in KPC Klebsiella pneumoniae bacteraemia: report of three cases. The Journal of antimicrobial chemotherapy. 2012 Nov:67(11):2777-9. doi: 10.1093/jac/dks270. Epub 2012 Jul 10
[PubMed PMID: 22782489]
Level 3 (low-level) evidence
[47]
Both A, Büttner H, Huang J, Perbandt M, Belmar Campos C, Christner M, Maurer FP, Kluge S, König C, Aepfelbacher M, Wichmann D, Rohde H. Emergence of ceftazidime/avibactam non-susceptibility in an MDR Klebsiella pneumoniae isolate. The Journal of antimicrobial chemotherapy. 2017 Sep 1:72(9):2483-2488. doi: 10.1093/jac/dkx179. Epub
[PubMed PMID: 28637339]
[48]
Hassoun-Kheir N, Hussien K, Karram M, Saffuri M, Badaan S, Peleg S, Aboelhega W, Warman S, Alon T, Pollak D, Szwarcwort Cohen M, Paul M. Clinical significance and burden of carbapenem-resistant Enterobacterales (CRE) colonization acquisition in hospitalized patients. Antimicrobial resistance and infection control. 2023 Nov 20:12(1):129. doi: 10.1186/s13756-023-01323-y. Epub 2023 Nov 20
[PubMed PMID: 37986092]
[49]
Mills JP, Marchaim D. Multidrug-Resistant Gram-Negative Bacteria: Infection Prevention and Control Update. Infectious disease clinics of North America. 2021 Dec:35(4):969-994. doi: 10.1016/j.idc.2021.08.001. Epub
[PubMed PMID: 34752228]
[50]
Gniadek TJ, Carroll KC, Simner PJ. Carbapenem-Resistant Non-Glucose-Fermenting Gram-Negative Bacilli: the Missing Piece to the Puzzle. Journal of clinical microbiology. 2016 Jul:54(7):1700-1710. doi: 10.1128/JCM.03264-15. Epub 2016 Feb 24
[PubMed PMID: 26912753]